Elevator Wire Rope, Lift Wire Rope, Elevator Wire Rope Design Realever Enterprise Limited , https://www.cnelevator-parts.com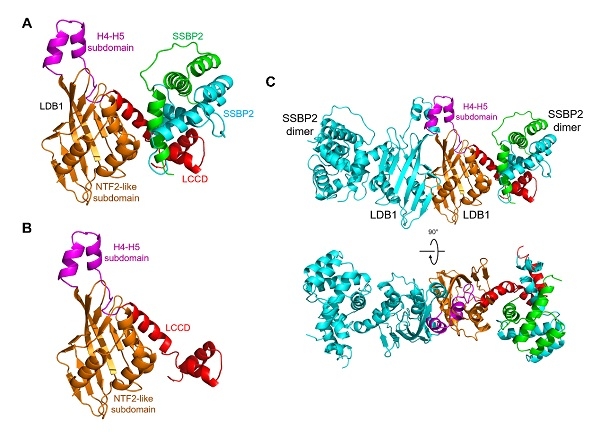
Enhancer control gene expression dimerizes and enhancer promoters interact
[ Instrument Network Instrument R & D ] On December 31, the Proceedings of the National Academy of Sciences (PNAS) published a paper entitled Crystal structure of human LDB1 in complex with SSBP2 online. This work was conducted by Xu Wenqing, Institute of Biophysics, Chinese Academy of Sciences / The team of Liang Dongcai and the Ann Dean team of the National Institutes of Health completed the cooperation.
Enhancers are small regions of DNA that can bind to proteins. After binding to proteins, the transcription of genes will be strengthened. Enhancers may be located upstream or downstream of the gene. It is not necessarily close to the gene to be acted on, or even on the same chromosome as the gene. This is because of the entangled structure of chromatin, so that positions far apart in the sequence also have the opportunity to touch each other.
Enhancers are switches that control the expression of genes. They are often far away from the genes they control and are located outside the coding frame. Therefore, DNA looping between the enhancer and the gene, which is far away from the gene, is a key step in gene transcription activation. In mammalian red blood cells, although the LDB1 protein does not bind DNA itself, it binds to the LDB1 transcription complex on long-range enhancers and erythrocyte-generating related genes, respectively. Long-distance enhancers and promoters can be achieved through the dimerization of LDB1. Interaction between them. The LDB1-SSBP complex is a core complex of many important protein complexes such as Wnt enhancer complex and LDB1 transcription complex, which is essential for development. The SSBP protein prevents LDB1 protein from being degraded in the 26S proteasome, maintains the stability of the LDB1 complex, and then promotes the assembly of the transcription complex and regulates gene expression. However, the molecular mechanism of how LDB1 interacts with SSBP and how LDB1 forms homodimers has not been revealed.
Enhancers can be divided into two types: cell-specific enhancers and inducible enhancers: â‘ tissue- and cell-specific enhancers. The enhancement effect of many enhancers is highly tissue-cell specific, and can only perform its function with the participation of specific transcription factors (proteins). â‘¡ inducible enhancer. The activity of this enhancer usually requires the participation of a specific promoter. For example, metallothionein genes can be transcribed in a variety of tissue cells, and can be induced by steroid hormones, zinc, cadmium, and growth factors to increase transcription levels.
This paper reports the crystal structure of the LDB1 / SSBP2 complex. The authors found that the LDB1 dimer domain (DD) contains an N-terminal nuclear transport factor 2 (NTF2) -like domain and a subdomain composed of α-helix 4 and α-helix 5, which together constitute the LDB1 dimer. surface. The two LDB / Chip conserved domains (LCCDs) of the LDB1 dimer are flanking the core DDs, and each LCCD interacts extensively with the SSBP2 dimer. The conserved linker between LDB1 DD and LCCD covers the potential ligand binding pocket of LDB1 NTF2-like subdomain, which may become the regulatory site of LDB1 structure and function. In addition, the structural and biochemical data in this article provide a structural basis for understanding how the LDB1 and LDB1 / SSBP2 interactions become structural cores of different complexes that regulate cell selection decisions and long-range enhancer-promoter interactions.
Promoter is a DNA sequence that RNA polymerase recognizes, binds and starts to transcribe. It contains the conserved sequences required for RNA polymerase specific binding and transcription initiation. Most of them are located upstream of the transcription initiation point of the structural gene. The promoter itself is not affected by Transcription. But some promoters (such as tRNA promoter) are located downstream of the transcription start point, and these DNA sequences can be transcribed. Promoter characteristics were originally identified by mutations that increased or decreased the rate of gene transcription. The promoter is generally located upstream of the transcription start site.